Researchers develop molecular markers for breeding of the rubber tree
Haikou, China – A research team from Chinese Academy of Tropical Agricultural Sciences has developed and validated a liquid single nucleotide polymorphism (SNP) chip named “HbGBTS80K,” which includes 80,080 SNPs evenly distributed across 18 chromosomes. This SNP chip effectively distinguished 404 rubber accessions into four groups in population genetic diversity analysis and detected the major gene HbPSK5 in GWAS for the number of laticifer rings. The HbGBTS80K chip is a valuable tool for accelerating functional studies and molecular breeding in rubber trees, addressing the inefficiencies of traditional breeding methods.
Molecular markers are specific DNA fragments that reflect differences among biological individuals, essential for marker-assisted breeding. While traditional markers like RFLP, RAPD, AFLP, and SSR have limited genome coverage, SNPs have become vital in plant genetic studies. Solid-phase SNP chips have advanced molecular breeding but face high costs and limitations in target loci fixation. Liquid-phase chips, like SNP-based GBTS, offer flexibility and cost-efficiency, yet are underutilized in rubber tree breeding.
A study published in Tropical Plants on 17 May 2024, aims to develop and validate a liquid SNP chip called “HbGBTS80K’ to enhance rubber tree breeding efficiency.
In this study, the HbGBTS80K chip was designed by first assembling the high-quality genome of the rubber tree cultivar “CATAS8-79” and re-sequencing 335 accessions at an average depth of ~20×. This process generated 5,323,701 SNPs, which were filtered based on minor allele frequency, deletion rate, and heterozygosity to select 96,044 SNPs for capture probes.
After evaluation with 69 additional accessions, 80,080 high-confidence SNP sites were retained. These SNPs were evenly distributed across the genome, with the highest density on chromosome 6 and the lowest on chromosome 1. Gene annotation revealed that 64.80% of SNPs were located in gene body region, including exonic, intronic, upstream, and downstream areas.
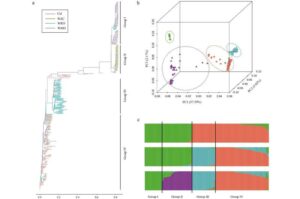
ADMIXTURE analysis using the HbGBTS80K chip classified 404 rubber accessions into four distinct groups, aligning with previous genetic diversity studies. The chip also demonstrated high accuracy in genome-wide association studies (GWAS) by identifying the major gene HbPSK5 for the number of laticifer rings (NLR), which is a key trait for natural rubber yield. This validation highlights the chip’s effectiveness in both genetic diversity analysis and functional gene identification, making it a valuable tool for advancing rubber tree molecular breeding.
According to the study’s senior researcher, Weimin Tian, “the HbGBTS80K liquid SNP chip is a valuable tool that will facilitate functional studies and molecular breeding of the rubber tree.”
In summary, the HbGBTS80K liquid SNP chip, developed from whole-genome resequencing of 335 rubber tree accessions, includes 80,080 SNPs distributed across 18 chromosomes. It effectively distinguishes rubber accessions into four groups and accurately identifies the major gene HbPSK5 associated with laticifer rings via GWAS. This chip is a valuable tool for advancing functional studies and molecular breeding of rubber trees. Looking ahead, the HbGBTS80K chip will facilitate the development of superior rubber tree varieties and serve as a model for similar advancements in other crops.